For the first time, a US scientific research team has drawn the 3D atomic structure of a key enzyme of Paramyxovirus, which can help develop antiviral drugs including anticoronavirus drugs. The research results were recently published in the Proceedings of the National Academy of Sciences. Paramyxovirus is a type of virus that includes common pathogens such as measles, mumps, human parainfluenza virus, and respiratory syncytial virus. Since both are RNA (ribonucleic acid) viruses, the mechanism of action of Paramyxovirus is similar to that of coronavirus. Enzymes play an important role in the assembly of RNA molecules and often become targets of antiviral drugs.
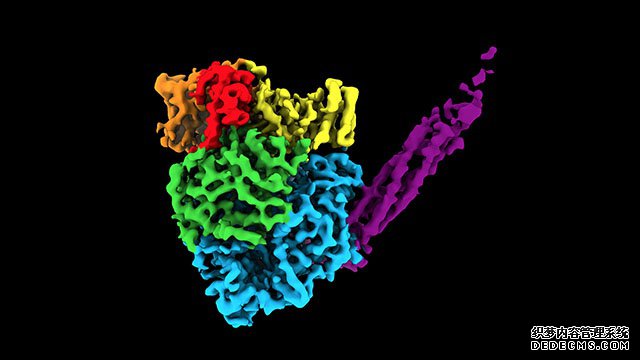
Researchers at Northwestern University used cryo-electron microscopy to observe the inside of the molecule, captured hundreds of thousands of 2D images of the human parainfluenza 5 polymerase, and used computer algorithms to reconstruct the 3D atomic structure of the enzyme.
The study found that this enzyme is an irregular sphere with a long tail composed of 4 phosphoproteins. The study also found that this virus uses the same protein to achieve genome replication and transcription.
Robert Lamb, a professor of molecular biology at Northwestern University in the United States, said that the traditional method of developing drugs is a bit of “luck”, hoping that drug candidates can hit the target. The clarification of the 3D atomic structure of the key enzymes of the virus is expected to be targeted when designing drugs. “We need more antiviral drugs to ensure that people can get treatment quickly when they are infected.”
(Editor in charge: admin)


0 Comments for “Research to map the 3D atomic structure of Paramyxovirus, or to help the development of antiviral drugs”